CalicoST - Inferring allele-specific copy number aberrations and tumor phylogeography from spatially resolved transcriptomics
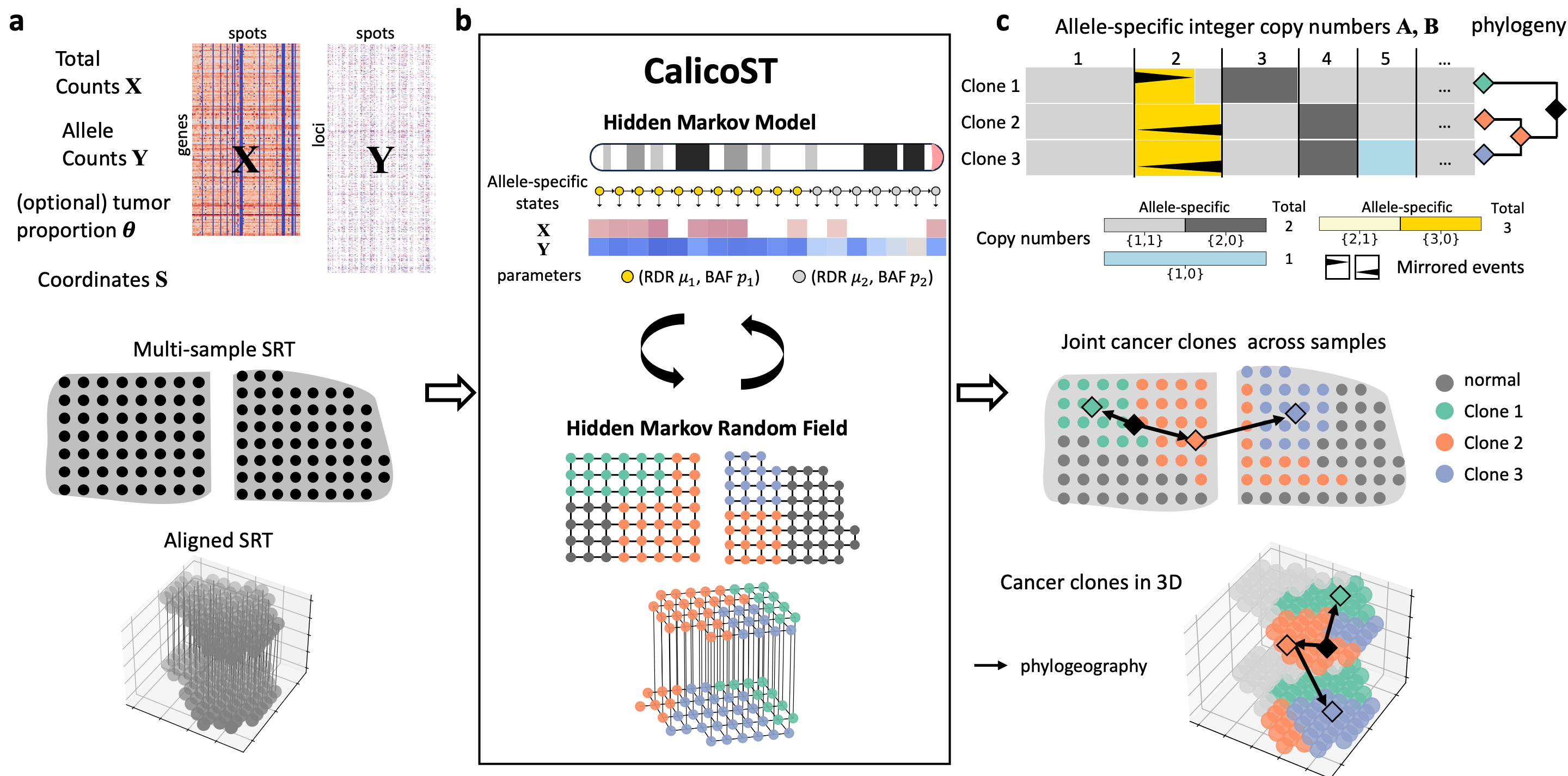
CalicoST is a probabilistic model that infers allele-specific copy number aberrations and tumor phylogeography from spatially resolved transcriptomics.CalicoST has the following key features: 1. Identifies allele-specific integer copy numbers for each transcribed region, revealing events such as copy neutral loss of heterozygosity (CNLOH) and mirrored subclonal CNAs that are invisible to total copy number analysis. 2. Assigns each spot a clone label indicating whether the spot is primarily normal cells or a cancer clone with aberration copy number profile. 3. Infers a phylogeny relating the identified cancer clones as well as a phylogeography that combines genetic evolution and spatial dissemination of clones. 4. Handles normal cell admixture in SRT technologies hat are not single-cell resolution (e.g. 10x Genomics Visium) to infer more accurate allele-specific copy numbers and cancer clones. 5. Simultaneously analyzes multiple regional or aligned SRT slices from the same tumor.
Installation
Find the details of installation here.
Getting started with CalicoST
Browse the Tutorials to get started with CalicoST here.